Mutations
Summary
Since the discovery of mutations by Hugo de Vries in 1901, the quest to understand the genetics behind cancer progression has occupied the interest of researchers worldwide. One of the key characteristics of cancer is genetic instability, meaning that there is a high frequency of deleterious mutations within the genome. These mutations may be the result of prolonged exposure to a carcinogen, such as smoking and ultraviolet (UV) radiation, or inheritance from a parent at the time of conception. Cancer’s ability to mutate and adapt to external stressors is one of the reasons why it is so difficult to treat. Databases like the Catalogue of Somatic Mutations in Cancer (COSMIC), the Cancer Genome Atlas (TCGA), and the International Cancer Genome Consortium (ICGC) have been essential in the identification of cancer-related mutations [1,2]. Building off of this wealth of knowledge, scientists are hard at work developing the best treatments and therapies to combat the sustained evolution of cancer mutations.

Kaylee Goodspeed
Undergraduate from Lamar University
The future of cancer treatment involves alternative regimens that target cancer at its source, with minimal damage to healthy surrounding tissues. This branch of precision oncology is a relatively recent development and is growing rapidly. Understanding the mechanisms behind biological components like specific pathway inhibitors, tumor suppressor genes, and biomarkers are paramount in making the connection between cancer etiology and treatment [3,4].
References
- Forbes, Simon A, et al. “COSMIC: Somatic Cancer Genetics at High Resolution.” Nucleic Acids Research 45 (2017): 777-783. doi:10.1093/nar/gkw1121
- Nakagawa, Hidewaki and Masashi Fujita. “Whole Genome Sequencing Analysis for Cancer Genomics and Precision Medicine.” Cancer Science (2018): 513-522. doi:10.1111/cas.13505
- Jette, Nicholas R., et al. “ATM-Deficient Cancers Provide New Opportunities for Precision Oncology.” Cancers 12 (2020): 1-13. doi:10.3390/cancers12030687
- Soldatos, Theodoros G., Sajo Kaduthanam, and David B. Jackson. “Precision Oncology – The Quest for Evidence.” Journal of Personalized Medicine 9 (2019): 1-17. doi:10.3390/jpm9030043
Full Article
The human body is the most sophisticated machine known to man. Every system is connected, and a malfunction in one area can upset the entire framework. A malady such as cancer is able to throw a wrench into the cogs of our metaphorical engines. Cancer is the unregulated division of cells that result in a malignant tumor. Many factors contribute to the development of a cancerous tumor, including genetic mutation, viral or bacterial infection, diet and lifestyle, and external influences. While most cases include a combination of the factors listed above, this paper will focus on the role of human genomic mutations in tumorigenesis. A mutation is a result of damaged genes by instances of DNA copying error, the inheritance of a faulty gene, or excessive exposure to a carcinogen (sunlight, radiation, smoking/asbestos). Research on tumor suppressor genes has allowed scientists to note that more than 50% of human cancers are characterized by mutations in the p53 gene, with the exception of hereditary mutations [1]. Further research is needed to predict, identify, and prevent the adaptation of cancer cells by mutation.
Imagine reading a good book. After becoming engrossed in the story and its characters, you stumble across a sentence that does not make any sense. There are misspelled words, and the order is all mixed up. You think nothing of it, attributing it to a possible printing error, and try to keep reading. Then suddenly, you notice that the whole paragraph does not belong. It mentions characters and events that have not even occurred yet! What in the world is going on? You flip through the rest of the book, hoping that the issue is not repeated. Alas, the remainder of the story is a jumbled mess of letters and entire chapters are missing! This sorrowful scenario is similar to how mutations occur.
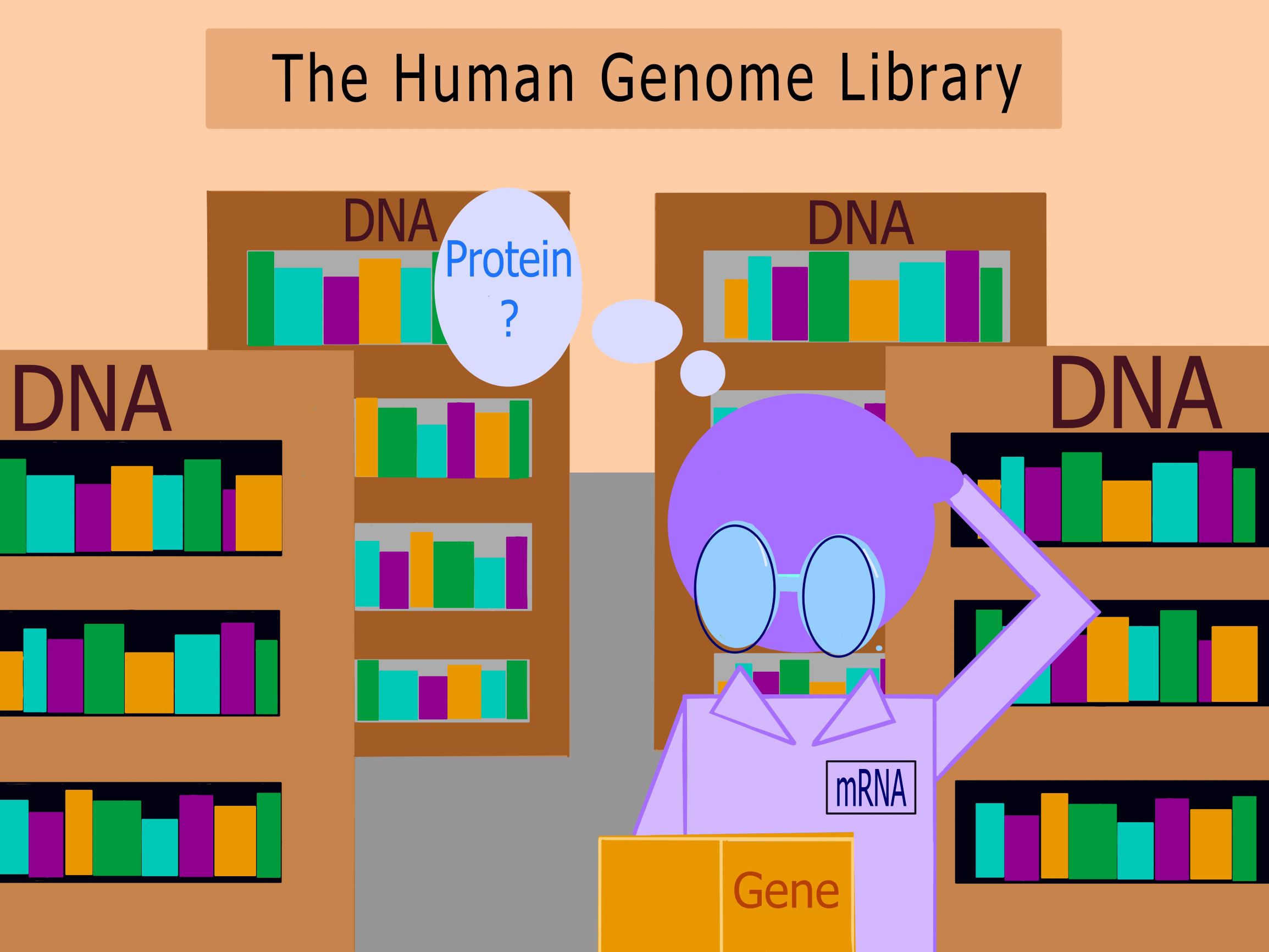
Figure 1. The Human Genome as a Library: The Human Genome can be represented by a library. The shelves (DNA) in the library house the knowledge the reader seeks. Each book (gene) is composed of words (nucleotides) to inform the reader (mRNA). If the words (nucleotides) are out of order, the reader (mRNA) cannot make sense of them. The reader’s understanding of the message (protein) portrayed by the book (gene) will be altered. When the reader (mRNA) goes to tell their friend about the book’s plot (protein), they will spread misinformation. Like in a game of Telephone, the incorrect message gets repeated down the chain until it has no resemblance to the original content.
The human genome can be described as our “biological library”, with each gene being a book within it. For a book to make sense, there has to be an order: words become sentences, sentences become paragraphs, paragraphs become chapters, and so on. The coding of proteins from our DNA acts in the same way (Figure 1). If there is a mutation in the genes of our DNA (shelves), the order of nucleotides arranged by mRNA (reader) will be faulty and may alter the function of the final protein. In some instances, the incessant replication of a faulty gene can lead to the development of cancer.
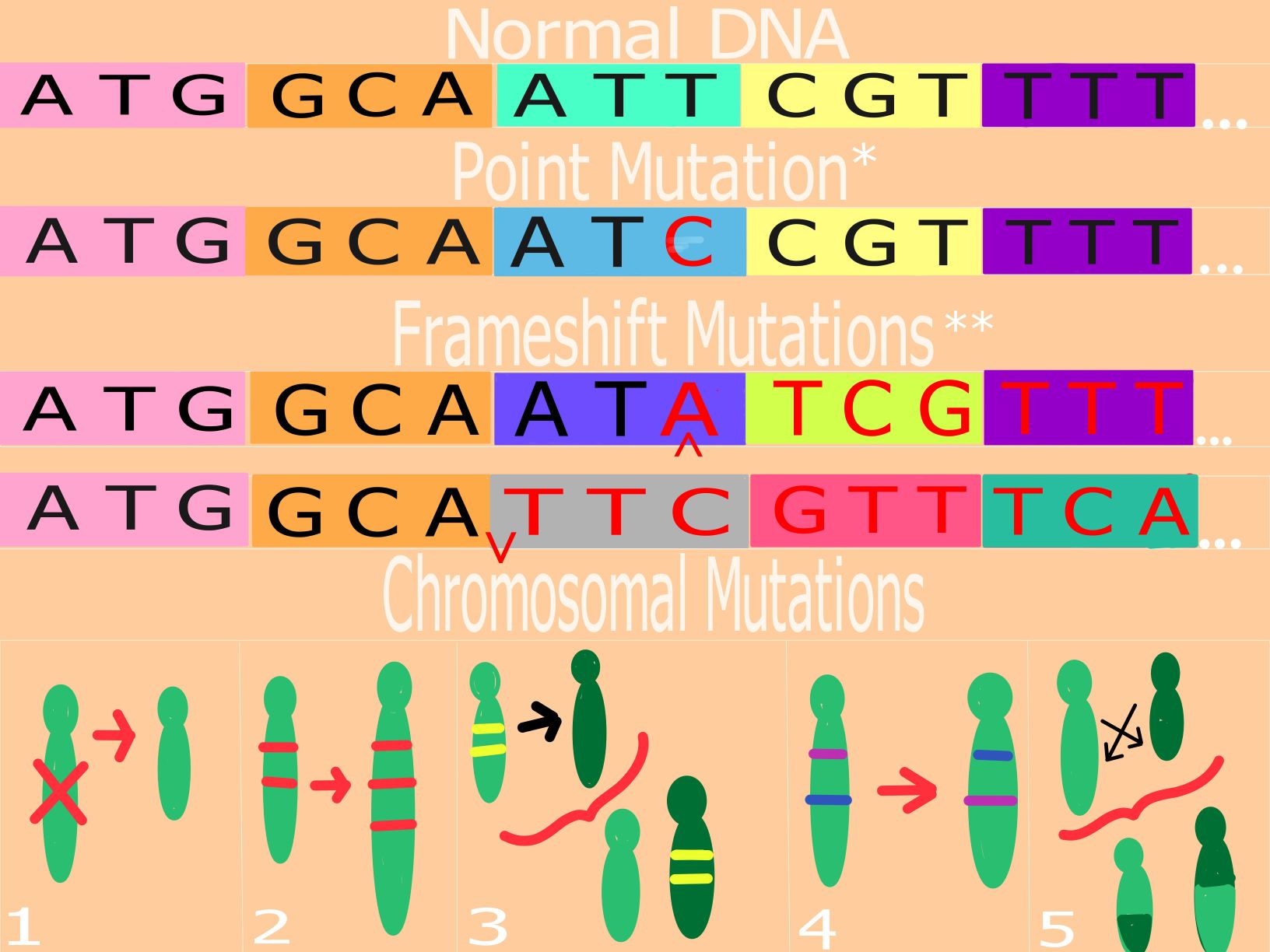
Figure 2. Common Somatic Mutations : Illustrations of the best-studied mutations that occur in the human genome, including point mutations, frameshift mutations (insertion and deletion), and chromosomal mutations.
Our DNA is made of chains of the four nucleotides: Adenine, Thymine, Guanine, and Cytosine. The nucleotides are further grouped into triplets called “codons”, which code for amino acids that build up to a protein. When mRNA “reads” our DNA, it creates a corresponding strand of RNA depending on what is translated from those codons. Sometimes mutations occur that affect the original message intended for the mRNA by our DNA. There are three well-understood somatic mutations that occur within the human genome. First are point mutations, where only one nucleotide is changed. There are three subcategories of point mutations: silent mutations, missense mutations, and nonsense mutations (Figure 2*). Silent mutations, as suggested by their name, do not change the amino acid coded and have no effect on the final protein. Missense mutations happen when a single nucleotide is swapped out for another, and that single amino acid will change. Nonsense mutations occur when the altered nucleotide results in the coding of a “stop codon”, effectively cutting off translation. Second to point mutations are frameshift mutations, of which there are two types (Figure 2**). Insertion mutations include the introduction of a nucleotide into the DNA sequence that shifts the nucleotides that follow and may also change the amino acids that those nucleotides originally coded for. Deletion mutations take place when a nucleotide is removed from the sequence, also shifting the following nucleotides and their amino acids. The third type of somatic mutation are chromosomal mutations.
There are five types of chromosomal mutations: deletion (1), duplication (2), insertion (3), inversion (4), and translocation (5) (Figure 2). A deletion chromosomal mutation occurs in a similar way to a frameshift deletion, except instead of the removal of a nucleotide, a section of the chromosome is removed. A duplication chromosomal mutation takes place when a section of the chromosome is doubled or amplified. Insertion chromosomal mutations are, once again, similar to a frameshift insertion except for the fact that a section of a chromosome is being removed instead of a nucleotide. An inversion chromosomal mutation happens when a section of the chromosome is flipped within the chromosome. Finally, a translocation chromosomal mutation occurs when a section from one chromosome is swapped with part of another chromosome.
The term mutation was first coined by Dutch botanist Hugo de Vries in 1901 after a discovery on his experiment with the evening primrose Oenothera lamarckiana. During the 20th century, the understanding of mutations was unclear due to differing perspectives ranging from Darwinism and Lamarckism to other biologists. Key to the discovery of mutations were the genetic tenets developed by Gregor Mendel. Mendel developed two laws, the Law of Segregation and the Law of Independent Assortment. The Law of Segregation states as chromosomes are separated in meiosis (sex cell division), each gamete (reproductive cell) only has one allele for a specific trait. In addition, the Law of Independent Assortment postulates that the inheritance of one gene does not determine that of another gene. After Mendel’s laws of genetics became known, more research was conducted to discover how mutations can lead to cancer. From this, researchers have learned that most cancers have resulted from multiple mutations in different genes and not just one. Studying the variety of mutations has led to the identification of specific mutations linked to certain cancers, like BRCA1 and BRCA2 mutations [2]. These mutations are known to be associated with genes that account for an estimated 25% of attributed cases of ovarian and breast cancer. Somatic mutations may differentiate into “driver” or “passenger” mutations. Whereas passenger mutations have no observed effect on tumor progression, driver mutations serve to promote tumor growth. Driver gene mutations involve three successive mutations, which are noted to have conferred growth advantage for cancer development [3]. The first one results in the creation of a benign lesion, which is a general term for anything non-cancerous and abnormal like an infected area or growth of skin. The second mutation results in another expansion of cells that can result in a benign tumor, which are non-cancerous abnormal masses of tissue and can be a lesion. The third driver gene mutation gives the tumor an ability to expand or metastasize and become malignant. Overall, these mutations promote tumorigenesis through a selective growth advantage and allow for additional clonal mutations [3]. Researchers have dedicated enormous effort toward identifying the mutations involved in cancer. The Catalogue of Somatic Mutations in Cancer (COSMIC), the world’s largest database for cancer mutations, began in 2004 with details of only four human genes [4]. Since its conception, COSMIC has accumulated information for more than 4 million human cancer mutations. Furthermore, scientists around the world have sequenced more than 50,000 cancer genomes, with The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) contributing 20,000 of that total [5].
The most effective treatments for mutation-driven cancers involve targeted therapies. Poly-ADP-ribose polymerase (PARP) inhibitors are particularly promising in the emerging field of precision oncology. PARP inhibitors were first discovered in 1971 when researchers observed their role in cell death, ischemia, and DNA repair [6]. In a process called “PARylation”, the PARP inhibitor binds to the ends of broken DNA strands and recruits nearby proteins to repair the damage [6]. In mouse models of breast cancer, the PARP inhibitor Olaparib showed antitumor activity and quickly moved into clinical trials. The FDA recently approved Olaparib for use in advanced cases of ovarian, breast, pancreatic, and prostate cancers [6]. Also, important to precision oncology are biomarkers that can be used to predict drug and therapy responses as well as potential mutations. The three broadest classes of biomarkers include prognostic biomarkers, predictive biomarkers, and pharmacodynamic biomarkers. Prognostic biomarkers can identify symptoms that are precursors to disease, and allow physicians to determine which therapies or treatments will aid best in halting the disease pathway; Predictive biomarkers measure the concentration of a drug at a specific site and make it easier to determine that drug’s efficacy; Pharmacodynamic biomarkers measure the relationship between a drug and its intended target. Because biomarkers sometimes predict conflicting outcomes, the traditional method of single-gene testing is becoming obsolete. Targeted gene panels are a potential solution to this predicament, as they provide higher sequencing depth, are more cost-effective, and are more efficient than other sequencing techniques [7]. Interpretation of these findings by a physician can aid in deciding which method of treatment would be most effective for the patient.
Mutations can be either hereditary or acquired, meaning a result of DNA copying error or excessive exposure to carcinogens. There are certain factors involved in hereditary mutations that an individual cannot control. While this is true, one can receive screenings to detect common gene mutations that are correlated to certain cancers to be proactive. As sequencing and inhibition techniques evolve, we look forward to future methods with the potential to halt cancer-causing mutations in their tracks.
References
- Basu AK. DNA Damage, Mutagenesis and Cancer. Int J Mol Sci. 2018;19(4):970. Published 2018 Mar 23. doi:10.3390/ijms19040970
- Hatano Y, Tamada M, Matsuo M, Hara A. Molecular Trajectory of BRCA1 and BRCA2 Mutations. Front Oncol. 2020;10:361. Published 2020 Mar 25. doi:10.3389/fonc.2020.00361
- Reiter JG, Baretti M, Gerold JM, et al. “An Analysis of Genetic Heterogeneity in Untreated Cancers.” Nat Rev Cancer. 2019;19(11):639-650. doi:10.1038/s41568-019-0185-x.
- Forbes, Simon A, et al. “COSMIC: Somatic Cancer Genetics at High Resolution.” Nucleic Acids Research 45 (2017): 777-783. doi:10.1093/nar/gkw1121
- Nakagawa, Hidewaki and Masashi Fujita. “Whole Genome Sequencing Analysis for Cancer Genomics and Precision Medicine.” Cancer Science (2018): 513-522. doi:10.1111/cas.13505
- Jette, Nicholas R., et al. “ATM-Deficient Cancers Provide New Opportunities for Precision Oncology.” Cancers 12 (2020): 1-13. doi:10.3390/cancers12030687
- Soldatos, Theodoros G., Sajo Kaduthanam, and David B. Jackson. “Precision Oncology – The Quest for Evidence.” Journal of Personalized Medicine 9 (2019): 1-17. doi:10.3390/jpm9030043